Micro and Molecular Biology
Deoxyribonucleic acid (DNA) is one of the most significant discoveries of the last century, primarily because it revolutionized many fields including genetics, molecular biology, medical research, agriculture, forensic science, and many others. DNA is a biological molecule that acts as the storage of genetic information essential for bodily function, growth, and the reproduction of all living organisms. This information is located in the nucleus of every cell in an organism and can be transmitted not only from one cell to another but also from parents to offspring. Hence, knowledge on the structure of DNA is very valuable in understanding how the transfer of genetic information occurs.
True Non-Contact Imaging of Various Samples
The need to measure sample surface features have become increasingly important in many applications such as manufacturing semiconductor devices and monitoring the structural characteristics of biological samples. There are some AFM techniques used in measurement of sample topography. Examples include tapping mode and contact mode. However, these modes require a tip-sample interaction that can lead to sample damage and tip quality degradation. The need for a technique that can measure surface topography without touching the sample surface is very real, especially for samples sensitive to surface deformation.
Cellulose Nanowhiskers Topography & Young’s Modulus Imaging Using Atomic Force Microscopy
Carbon nanotubes (CNTs) are a popular sample for various nanotechnology studies. However, they are very difficult to synthesize en masse. As a result, researchers have started looking at alternatives with similar properties. One of the candidates to replace CNTs in some nanocomposite materials is cellulose nanowhiskers (CNWs), structures that can be readily produced from plant sources.
Hair Damage From Sunlight Radiation Characterized Using Atomic Force Microscopy
Products for sun protection such as hats, sunscreen, and clothing are commonly used to prevent skin irritation and damage, but few products for protecting hair exist. Through researching the radiation effects of sunlight on hair, it is to be determined whether or not this is a concern worth looking into, especially for those with less photostable, fair-colored hair. With the Park NX20 Atomic Force Microscope (AFM) and its user-friendly Park SmartScan software’s Auto mode, images of hair can be produced to compare its nanoscale topography before and after prolonged exposure to sunlight.
Single Strand DNA Molecules
NC-AFM Imaging of Bio-molecular Samples
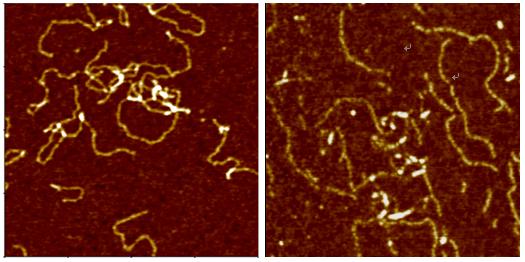
Deoxyribonucleic Acid (DNA) is a polymer chain that stores hereditary information of species. It is made from repeating units called nucleotides. In its natural state, the chain is 2.2 nm ~ 2.6 nm, and one nucleotide unit is 0.33 nm long. Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. While X-ray crystallography and electron microscopy are the main tools used to resolve the structure of DNA in 3-dimensions, scanning probe techniques have gradually gained importance in the field of high resolution DNA imaging due to their ease of sample preparation (in-necessity to crystallize the DNA prior to imaging) as well as the possibility of functional studies (e.g., study of protein–DNA interaction processes) under physiological conditions. In preparation for AFM imaging, DNA samples are usually immobilized onto a stiff, flat substrate surface. The negatively charged backbone of the DNA can be utilized for immobilization onto charged substrates such as freshly cleaved mica via electrostatic interactions. After the DNA molecules are adsorbed onto the substrate, the surface can be washed, air-dried, and promptly imaged. Alternatively, the imaging process can be carried out in physiological solutions.
Publications Using the XE-AFM on DNA Molecules
Journal: Nucleic Acids Research, 33(20) November 2005. 6515.
Title: Synthesis of novel poly(dG)–poly(dG)–poly(dC) triplex structure by Klenow exofragment of DNA polymerase I
Authors: A. Kotlyar, N. Borovok1, T. Molotsky1, D. Klinov, B. Dwir and E. Kapon;
System: Park Systems XE-100
Abstract: The extension of the G-strand of long (700 bp) poly(dG)–poly(dC) by the Klenow exo-fragment of DNA polymerase I yields a complete triplex structure of the H-DNA type. Atomic force microscopy morphology imaging shows that the synthesized structures lack single-stranded fragments and have approximately the same length as the parent 700 bp poly(dG)–poly(dC). CD spectrum of the polymer has a large negative peak at 278 nm, which is characteristic of the poly(dG)–poly(dG)–poly(dC) triplex. The polymer is resistant to DNase and interacts much more weakly with ethidium bromide as compared with the double-stranded DNA.
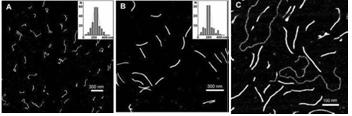
Fig. 2. AFM images of (A) synthesized poly(dG–dG)–poly(dC) molecules , (B) ~700 bp parent poly(dG)–poly(dC) and (C) poly(dG–dG)–poly(dC) molecules co-deposited with a linear plasmid pSK+ DNA on mica. Statistical length analyses of single, well-separated molecules from several images at several areas are shown in the inserts to (A) and (B).




